The gene search is a tool for searching genes in SGN's database. These genes originate from various sources, including GenBank, PubMed literature mining, historic collections of gene names, and SGN user-contributed data.
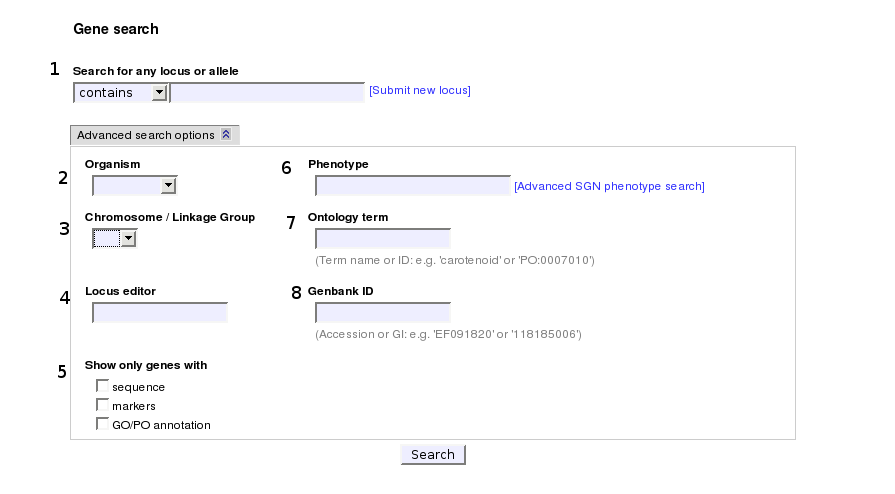
1. Simple search option: search by locus or allele name or symbol
Use this option for searching the database for genes with a locus or allele name/symbol/synonym/description containing what you typed in. You can search these fields for an exact match, begin with or end with your input, by selecting the desired option from the drop-down menu. The search is case-insensitive.
The following search options can be combined with the generic locus/allele search option. These are visible by clicking the 'Advanced search options' button:.
2. Organism
In this drop-down menu you will see a list of the common names of species that make up the collection of genes. You can use this optionfor limiting your search results to loci of only the organism you selected.
3. Chromosome
Choose a chromosome from this menu to limit results to loci mapped on that chromosome or linkage group.
4. Locus editor
Each locus in the genes database has an owner (either an SGN curator or a submitter). the owner also has certain edit privileges. You can search for loci owned by a specific SGN user by typing a first or a last name. Anyone interested in obtaining an SGN user account may do so by signing up for a new account. After obtaining an SGN user account you may upload new loci to SGN's database. If you are interested in obtaining editor privileges to a specific locus, please follow the link in the locus details page.
5. Annotation features
Use these checkboxes for narrowing your search to only genes with one or more of the following features:
- Sequence: genes that have sequence annotation (GenBank ID and/or SGN unigene ID)
- Marker: genes that correspond to a specific marker from SGN's marker database
- GO/PO annotation: genes that are annotated to at least one Gene Ontology or Plant Ontology term
6. Phenotype
Some genes may have a phenotype description, either as an associated allele, or an associated phenotyped accession. This option will narrow the search results for only genes with a phenotype description containing the word or string you typed in. For searching phenotypes of accessions not necessarily associated with a gene, you should use SGN's phenotype search page.
7. Ontology term
Use this box for searching for genes that are annotated to specific Gene Ontology or Plant Ontology terms. You may search for ontology annotations by typing any part of the ontology term ID, term name, term synonym, or term definition.
8. GenBank ID
Use this box for searching genes annotated to a specific GenBank ID. You may search using either a GI number or a GenBank accession.